
|
||
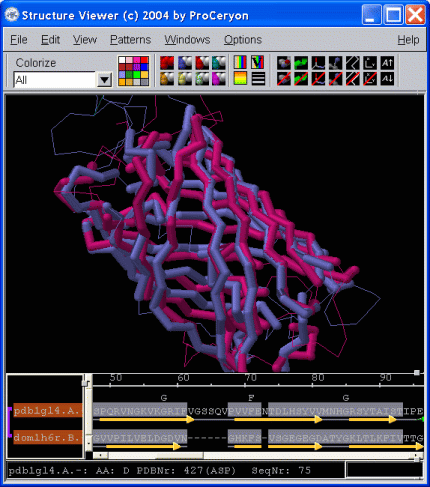
The Structure Viewer is specifically designed to investigate alignments and 3D structures simulteanously. The tight coupling of alignments and 3D models enables the researcher to quickly recognize conserved features shared by the aligned proteins. Key residues, active sites, ligand binding sites, or sequence patterns may be localized in the alignment as well as in the 3D structure. If installed on your system, sequence-to-structure alignments may be sent to the modeling package Modeller to perform automated side and loop modeling. |
||
| ||
|
|
|
| ||

|
|
|
| ||

|
|
|